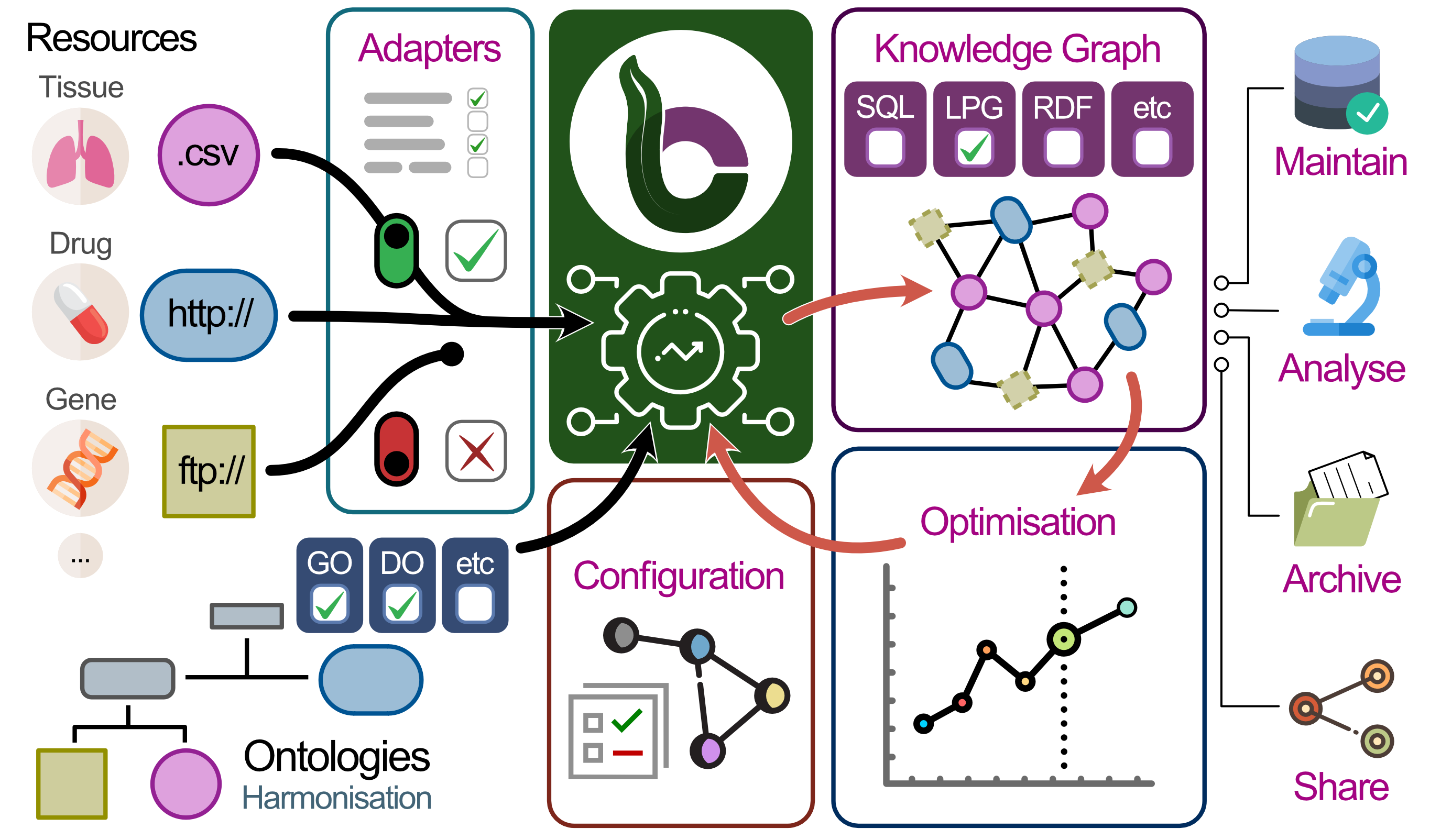
The Biocypher project addresses a broad collection of problems of annotation and interpretation of genomic data:
The mission statement is:
We aim to enable access to versatile and powerful knowledge graphs for as many researchers as possible. Making biomedical knowledge “their own” is often a privilege of the companies and groups that can afford individuals or teams working on knowledge representation in a dedicated manner. With BioCypher, we aim to change that. To achieve this, we have developed a framework that facilitates the creation of knowledge graphs that are informed by the latest developments in the field of biomedical knowledge representation. However, to make this framework truly accessible and comprehensive, we need the input of the biomedical community. We are therefore inviting you to join us in this endeavour!
This package, biocBiocypher, explores relationships between Biocypher and Bioconductor.
The initial work involves reflecting processes reviewed in tutorials into R, via basilisk and reticulate.
Here’s an illustration of how the IgA nephropathy knowledge base can be explored:
# assumes https://github.com/biocypher/igan.git README instructions
# followed, docker compose up -d succeeded and localhost:7474 gives
# a neo4j browser interface (at localhost:7687/browser on my machine)
#
# BiocManager::install("neo4j-rstats/neo4r")
# want 0.1.3
library(neo4r)
library(magrittr)
con <- neo4j_api$new(
url = "http://localhost:7474", user="", password="")
con$ping() # not 404!
'MATCH (n:Disease) RETURN n LIMIT 25;' %>% call_neo4j(con)