Basic concepts
Introduction to biocViews
“biocViews” is the name of a package in Bioconductor that performs many roles
- it was initially proposed to emulate CRAN task views, but did not achieve the levels of curation and narration present there
- it was implemented as an instance of Bioconductor’s “graph” class
- every vocabulary element is node in the biocViews graph
- “is a” conceptual relationships are expressed using directed edges
Illustration using R:
## A graphNEL graph with directed edges
## Number of Nodes = 497
## Number of Edges = 496## [1] "BiocViews" "Software" "AnnotationData"
## [4] "ExperimentData" "Workflow" "AssayDomain"
## [7] "Technology" "ResearchField" "BiologicalQuestion"
## [10] "WorkflowStep" "WorkflowManagement" "StatisticalMethod"## $BiocViews
## [1] "Software" "AnnotationData" "ExperimentData" "Workflow"
##
## $Software
## [1] "AssayDomain" "Technology" "ResearchField"
## [4] "BiologicalQuestion" "WorkflowStep" "WorkflowManagement"
## [7] "StatisticalMethod" "Infrastructure" "ShinyApps"
##
## $AnnotationData
## [1] "Organism" "ChipManufacturer" "CustomCDF"
## [4] "CustomArray" "CustomDBSchema" "FunctionalAnnotation"
## [7] "SequenceAnnotation" "ChipName" "PackageType"Quick view of EDAM in R
The following has a one-time startup cost of introducing infrastructure for working with owlready2 (python) in R.
## Loading required package: ontologyIndex
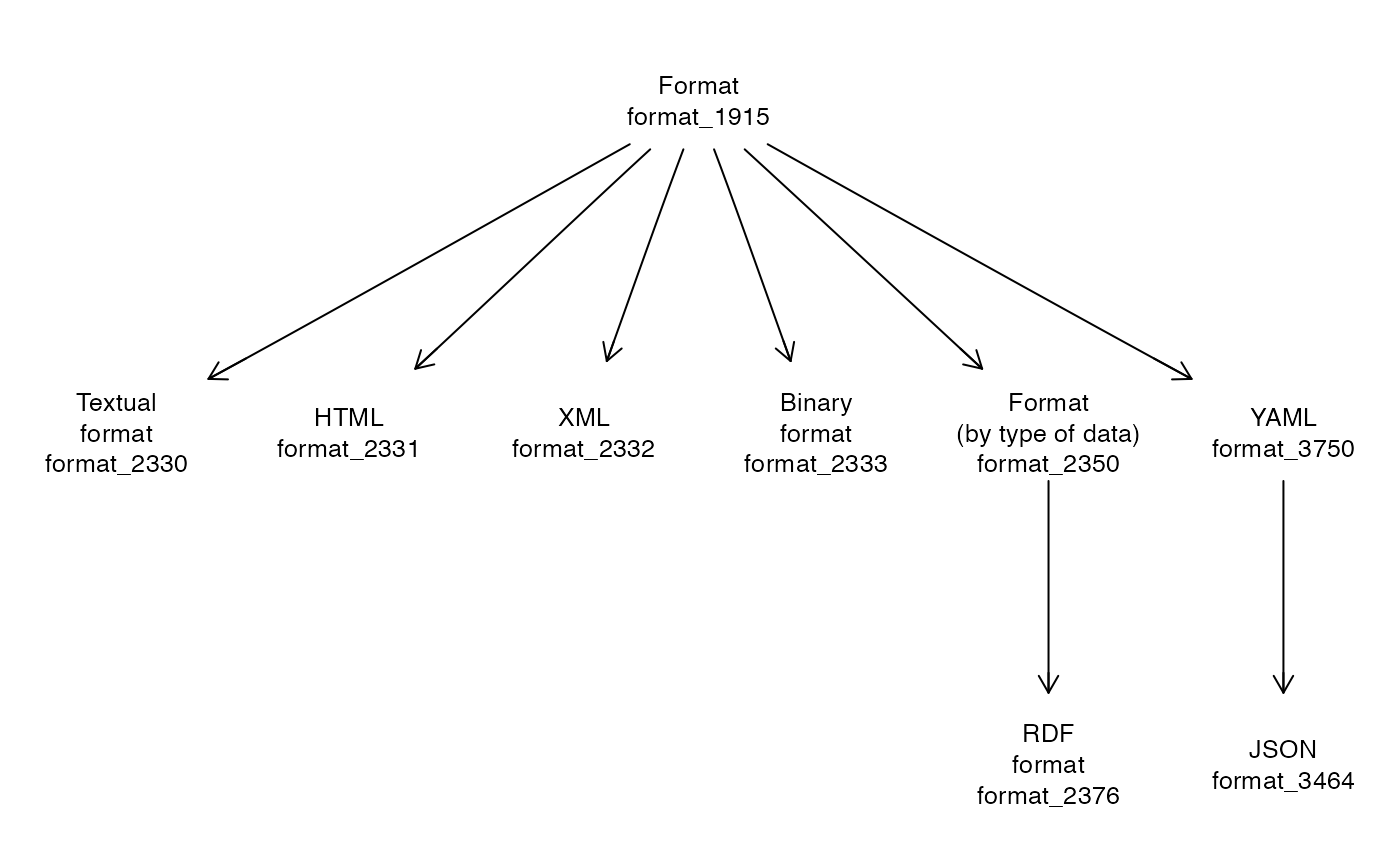
ed = setup_entities2("https://edamontology.org/EDAM_1.25.owl")
head(ed$name, 12)## data_0006 format_1915 operation_0004 data_0842
## "Data" "Format" "Operation" "Identifier"
## topic_0003 data_0005 DeprecatedClass data_2337
## "Topic" "Resource type" NA "Resource metadata"
## data_0007 data_0958 data_0581 data_0957
## "Tool" "Tool metadata" "Database" "Database metadata"
onto_plot2(ed, names(ed$name[ed$children$format_1915]))