bbsBuildArtifacts: filtering build reports
Vincent J. Carey, stvjc at channing.harvard.edu
July 16, 2023
Source:vignettes/bbsBuildArtifacts.Rmd
bbsBuildArtifacts.RmdIntroduction
We’d like to be able to get a quick overview of status for a subset of Bioconductor packages. “Status” is relative to Bioconductor version, package version, build platform, and condition of the platform.
We want to be able to work with the current artifacts provided at, e.g., https://bioconductor.org/checkResults/3.17/bioc-LATEST/report.tgz. Such gzipped tar resources are prepared for different types of resource.
suppressMessages({
suppressPackageStartupMessages({
library(bbsBuildArtifacts)
library(dplyr)
library(DT)
library(ggplot2)
})
})
bbsBuildArtifacts:::valid_types()## [1] "bioc" "data-experiment" "workflows" "books"
## [5] "bioc-longtests"We’ll focus on type bioc for now, which associates with Software packages. We don’t yet know if report.gz has the same structure for all types, but we hope so.
Our objective is to learn the status and processing times for various phases of the build process for all packages, and to analyze error, warning and note events programatically.
Artifact set manager
We define an S4 class ArtifSet to manage key information about builds. ArtifSet instances are produced using setup_artifacts.
We’ve produced a thinned version of the BBS report.tgz accessible in the package at demo_path().
od = getwd()
td = tempdir()
setwd(td)
untar(demo_path())
af = setup_artifacts(type="bioc", version="3.17", extracted=".")
af## bbsBuildArtifacts ArtifSet instance.
## 11 pkg paths for type bioc, Bioconductor version 3.17.
## 18 extra file paths.
## tarball production date: 2022-09-09
## R version: 4.2.1 (2022-06-23) -- "Funny-Looking Kid"
## Platforms:
## Linux (Ubuntu 20.04.4 LTS)
## macOS 10.14.6 Mojave
## Windows Server 2022 Datacenter
## Use paths(aset)[...] to retrieve selected paths.Information of immediate interest can be derived and tabulated in a data.frame instance.
setwd(td)
d = as.data.frame(af)
dim(d)## [1] 132 6
head(d)## host pkgname pkgversion status elapsed_time phase
## 1 nebbiolo1 flowMerge 2.44.0 OK 18.1 install
## 2 nebbiolo1 flowMerge 2.44.0 OK 53.2 buildsrc
## 3 nebbiolo1 flowMerge 2.44.0 OK 90.3 checksrc
## 4 nebbiolo1 flowMerge 2.44.0 NA NA buildbin
## 5 nebbiolo1 frma 1.48.0 OK 25.2 install
## 6 nebbiolo1 frma 1.48.0 OK 59.1 buildsrcWe use this to get statistics about timings for package installation, building and checking.
## merida1 nebbiolo1 palomino3
## 1.4291111 0.7262222 0.9954444## buildbin buildsrc checksrc install
## 0.1893056 0.8889444 1.8265833 0.2459444
ggplot(mutate(d, elapsed_time_sec=elapsed_time), aes(y=elapsed_time_sec, x=host)) +
geom_boxplot() + facet_grid(.~phase) + scale_y_log10() ## Warning: Removed 11 rows containing non-finite values
## (`stat_boxplot()`).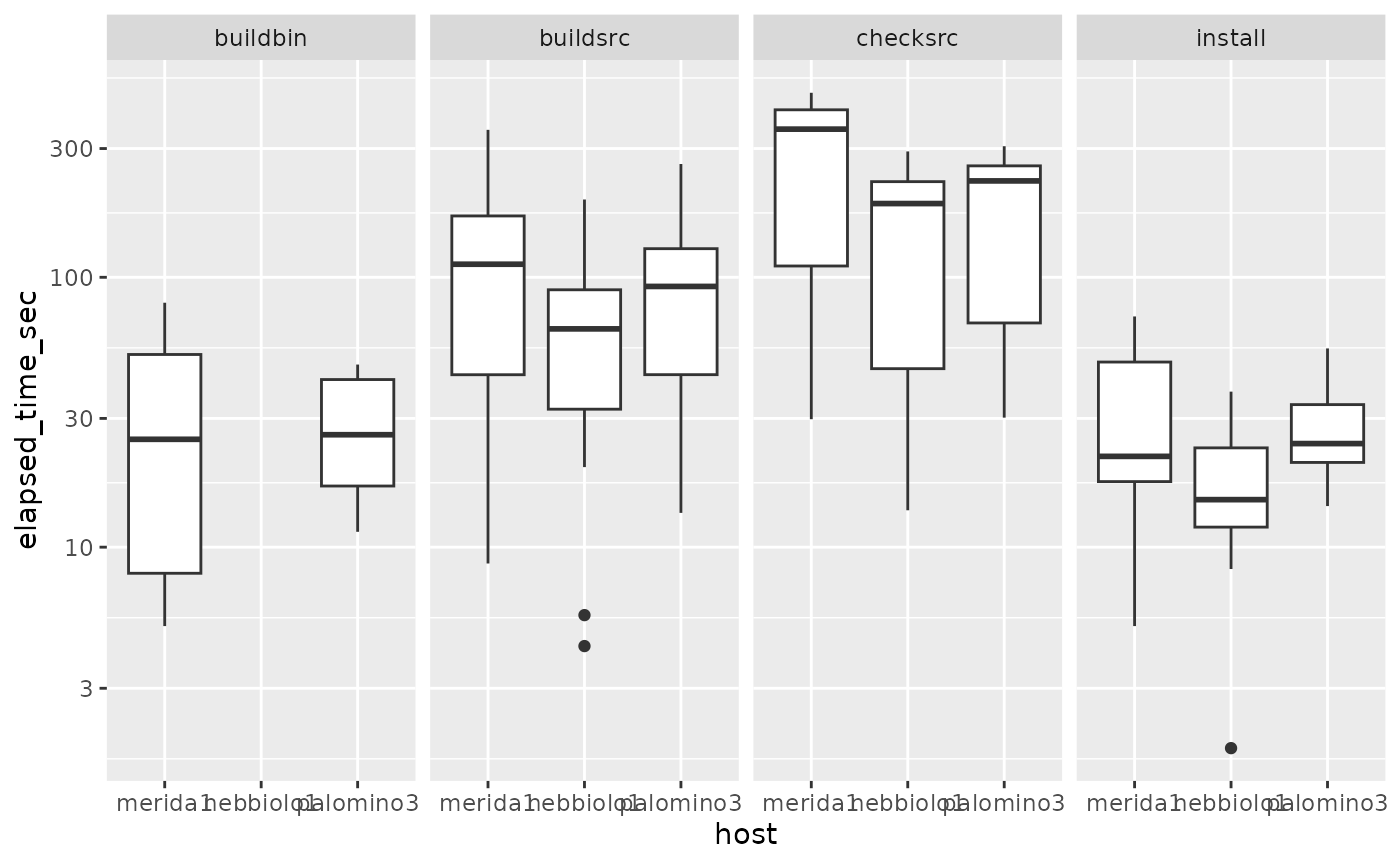
Information recorded about a package
Because the build/check processes are error prone, some packages may lack information described below.
raw_info
This is a selection from content of an info.dcf produced for each package.
## Formal class 'BBS_raw_pkg_info' [package "bbsBuildArtifacts"] with 6 slots
## ..@ name : chr "zinbwave"
## ..@ last_commit_date: POSIXct[1:1], format: "2022-04-26 15:41:22"
## ..@ version :List of 1
## .. ..$ Version:Classes 'package_version', 'numeric_version' hidden list of 1
## .. .. ..$ : int [1:3] 1 18 0
## .. ..- attr(*, "class")= chr [1:2] "package_version" "numeric_version"
## ..@ commit_tag : Named chr "678c0f6"
## .. ..- attr(*, "names")= chr "git_last_commit"
## ..@ branch : Named chr "RELEASE_3_15"
## .. ..- attr(*, "names")= chr "git_branch"
## ..@ maint_email : Named chr "risso.davide at gmail.com"
## .. ..- attr(*, "names")= chr "MaintainerEmail"Detailed information about events
Here’s how we can obtain the installation log for SummarizedExperiment on nebbiolo2.
setwd(td)
pd1 <- make_BBS_package_data(af, "zinbwave")
pd1## BBS_package_data for package 'zinbwave' version 3.17
names(slot(pd1, "host_data"))## [1] "nebbiolo1" "merida1" "palomino3"
hd = slot(pd1, "host_data")
cat(slot(hd$nebbiolo1, "install"), sep="\n")## ##############################################################################
## ##############################################################################
## ###
## ### Running command:
## ###
## ### /home/biocbuild/bbs-3.15-bioc/R/bin/R CMD INSTALL zinbwave
## ###
## ##############################################################################
## ##############################################################################
##
##
## * installing to library ‘/home/biocbuild/bbs-3.15-bioc/R/library’
## * installing *source* package ‘zinbwave’ ...
## ** using staged installation
## ** R
## ** data
## *** moving datasets to lazyload DB
## ** inst
## ** byte-compile and prepare package for lazy loading
## ** help
## *** installing help indices
## ** building package indices
## ** installing vignettes
## ** testing if installed package can be loaded from temporary location
## ** testing if installed package can be loaded from final location
## ** testing if installed package keeps a record of temporary installation path
## * DONE (zinbwave)Much of the content of these logs is routine unilluminating chatter. Isolating the information of functional value is a project for future effort.