From the primary site opencravat.org:
OpenCRAVAT is distinguished from similar tools by the amount and diversity of data resources and computational prediction methods available, which span germline, somatic, common, rare, coding and non-coding variants. We have designed the OpenCRAVAT resource catalog to be open and modular to maximize community and developer involvement, and as a result the catalog is being actively developed and growing larger every month.
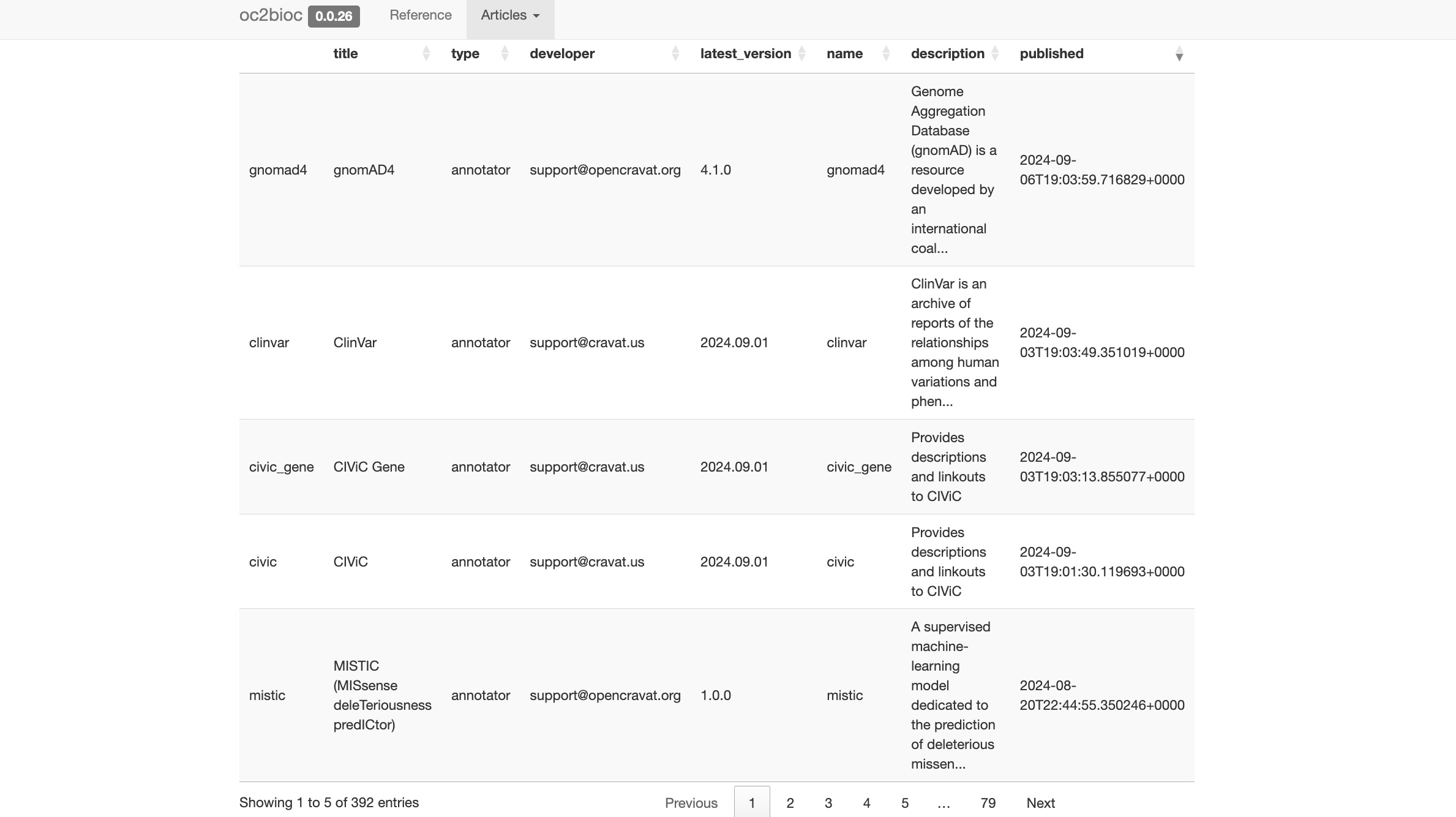
This package includes an R function to enumerate available annotators by querying the OpenCRAVAT REST API. Some recent versions:
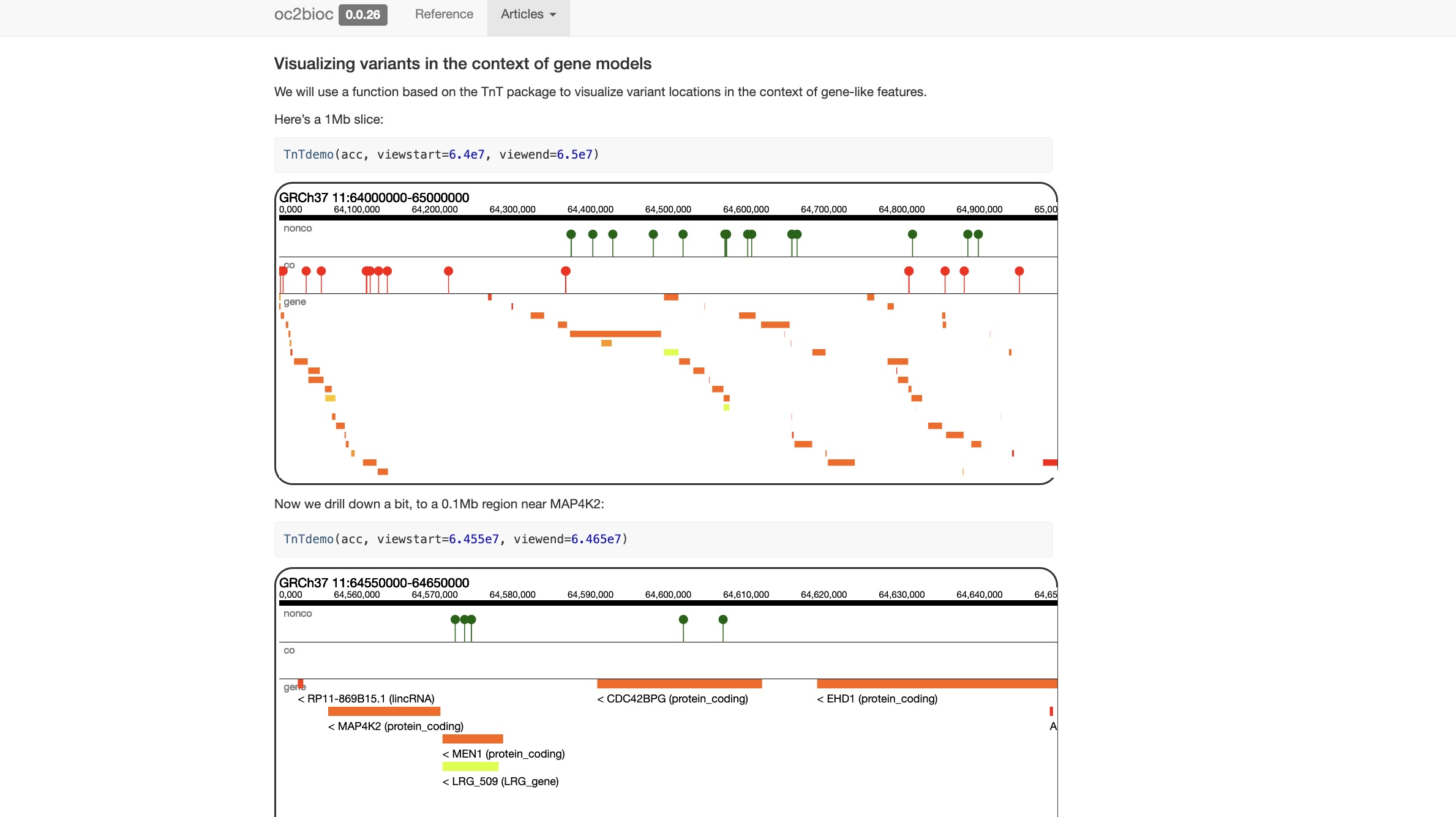
See the vignette for code that produces interactive visualization of identified variants: