Ontology concepts and tools in Bioconductor for Bioc2024
Vincent J. Carey, stvjc at channing.harvard.edu
August 10, 2025
Source:vignettes/owlents.Rmd
owlents.RmdObjectives
introduction to ontology-related computations in R with the ORDO rare disease ontology and CL cell ontology
review of general ontology resources and the OWL RDF/XML serialization
retrieving, updating, searching for OWL representations of ontologies using
BiocFileCacheandowl2cacheOntology mapping of phenotypes of the GWAS catalog using Experimental Feature Ontology
Introduction
ORDO for rare diseases
Our first example uses the ORDO ontology, which is distributed in compressed OWL format with the ontoProc package.
Here are the first 18 lines of the OWL XML:
library(ontoProc)
pa = get_ordo_owl_path()
readLines(pa, 18)## [1] "<?xml version=\"1.0\"?>"
## [2] "<rdf:RDF xmlns=\"http://www.w3.org/2002/07/owl#\""
## [3] " xml:base=\"http://www.w3.org/2002/07/owl\""
## [4] " xmlns:dc=\"http://purl.org/dc/elements/1.1/\""
## [5] " xmlns:efo=\"http://www.ebi.ac.uk/efo/\""
## [6] " xmlns:obo=\"http://purl.obolibrary.org/obo/\""
## [7] " xmlns:owl=\"http://www.w3.org/2002/07/owl#\""
## [8] " xmlns:rdf=\"http://www.w3.org/1999/02/22-rdf-syntax-ns#\""
## [9] " xmlns:xml=\"http://www.w3.org/XML/1998/namespace\""
## [10] " xmlns:xsd=\"http://www.w3.org/2001/XMLSchema#\""
## [11] " xmlns:ORDO=\"http://www.orpha.net/ORDO/\""
## [12] " xmlns:rdfs=\"http://www.w3.org/2000/01/rdf-schema#\""
## [13] " xmlns:skos=\"http://www.w3.org/2004/02/skos/core#\""
## [14] " xmlns:terms=\"http://purl.org/dc/terms/\""
## [15] " xmlns:licenses=\"https://creativecommons.org/licenses/\""
## [16] " xmlns:oboInOwl=\"http://www.geneontology.org/formats/oboInOwl#\">"
## [17] " <Ontology rdf:about=\"https://www.orphadata.com/data/ontologies/ordo/last_version/ORDO_en_4.2.owl\">"
## [18] " <versionIRI rdf:resource=\"https://www.orphadata.com/data/ontologies/ordo/last_version/ORDO_en_4.2.owl\"/>"This establishes a variety of namespaces and other metadata for annotations that will be used throughout the ontology. Of particular interest is the version string towards the end. As of 3 July 2024, version 4.5 has been published. We are working with version 4.2.
ontoProc uses an interface (via the reticulate package) to the owlready2 python modules distributed via pypi.
Ingestion is typically fast and returns an instance of the S3 class owlents.
orde = setup_entities(pa)
orde## owlents instance with 15239 classes.The most common conceptual relationship encoded in the ontologies to be considered is “is-a.” Given any term in the ontology, its ancestors are terms that are successively more general.
Let’s look at two terms. Given an owlents instance x and a numeric vector n, x[n] ‘subsets’ the information to the selected terms.
orde[1000:1001]## owlents instance with 2 classes.
ancestors(orde[1000:1001])## $Orphanet_79083
## {ORDO.Orphanet_79083, owl.Thing, ORDO.Orphanet_377788, ORDO.Orphanet_C001, ORDO.Orphanet_557493}
##
## $Orphanet_146
## {owl.Thing, ORDO.Orphanet_377788, ORDO.Orphanet_C001, ORDO.Orphanet_146, ORDO.Orphanet_557493}
labels(orde[1000:1001])## Orphanet_79083
## "PPARG-related familial partial lipodystrophy"
## Orphanet_146
## "Differentiated thyroid carcinoma"Here we are seeing a mix of R and python references. As this package matures, this mixing should diminish.
We can use character tags to subset owlents instances as well. This is how we “decode” two of the ancestors of orde[1000].
## Orphanet_377788 Orphanet_557493
## "Disease" "disorder"A new feature of ontoProc’s OWL processing is the availability of searching labels with regular expression matching. This feature is built in to owlready2.
lsrch = search_labels(orde$owlfn, "*lipodystrophy*")
length(lsrch)## [1] 26To visualize relationships among the terms, a plot method leverages the ontologyIndex packages from CRAN.
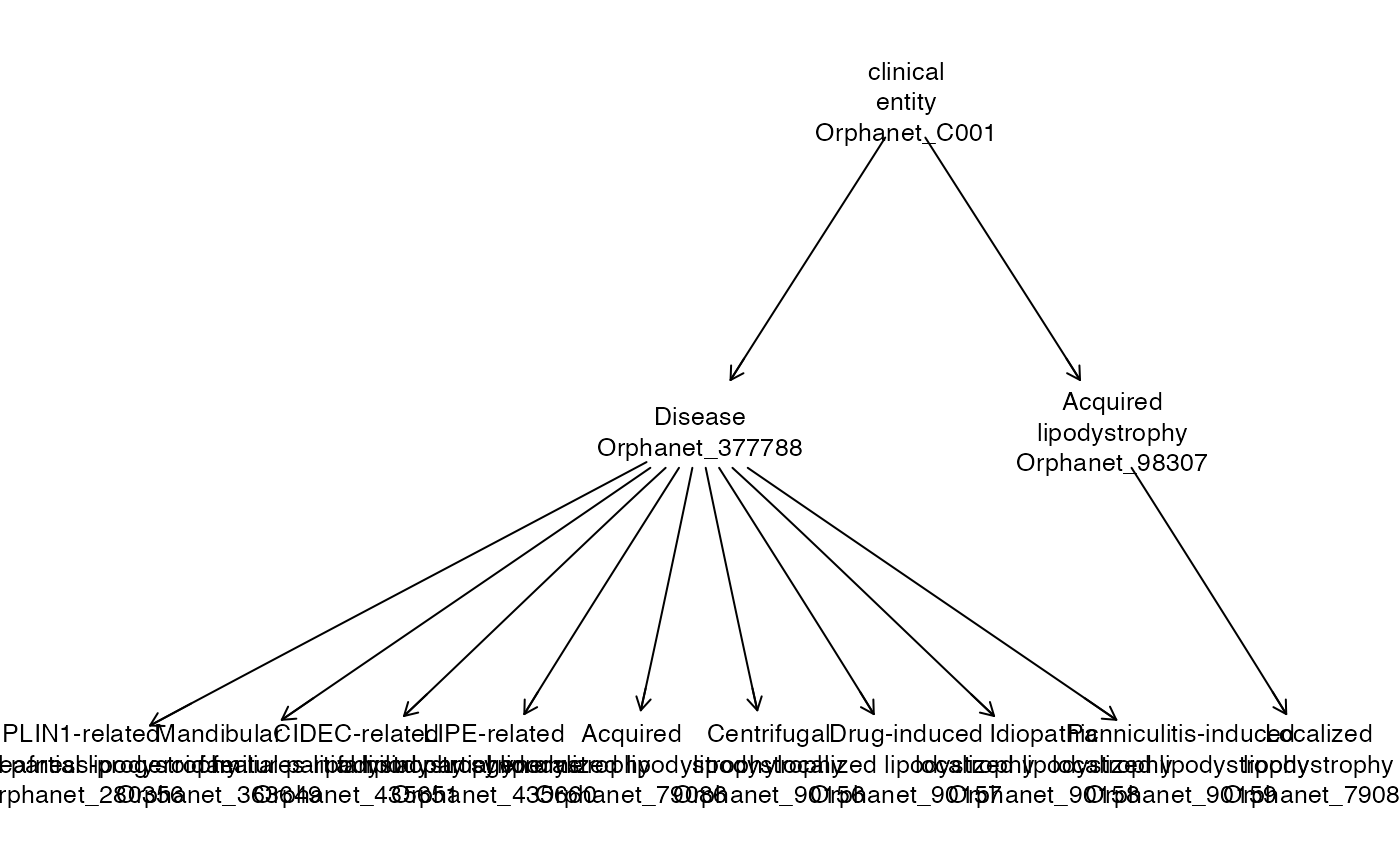
Cell ontology
The owl2cache function retrieves OWL from a URL or file and places it in a cache to avoid repetitious retrievals. The default cache is the one defined by BiocFileCache::BiocFileCache(). Here we work with the cell ontology. setup_entities will use owlready2 python modules to parse the OWL and produce an instance of S3 class owlents.
library(ontoProc)
clont_path = owl2cache(url="http://purl.obolibrary.org/obo/cl.owl")
# need to fix!
tmp = readLines(clont_path)
bad = grep("STATO_0000416", tmp)[1:2] # see https://github.com/obophenotype/cell-ontology/issues/3237
tmp = tmp[-bad]
bad = grep("STATO_0000663", tmp)[1:2] # see https://github.com/obophenotype/cell-ontology/issues/3237
tmp = tmp[-bad]
tf = tempfile()
writeLines(tmp, tf)
cle = setup_entities(tf)
cle## owlents instance with 18718 classes.A plot method is available. Given a vector of tags as reported in OWL (no colons are used), the plot method produces an ontologyIndex instance and runs onto_plot2 on the result.
sel = c("CL_0000492", "CL_0001054", "CL_0000236",
"CL_0000625", "CL_0000576",
"CL_0000623", "CL_0000451", "CL_0000556")
plot(cle, sel)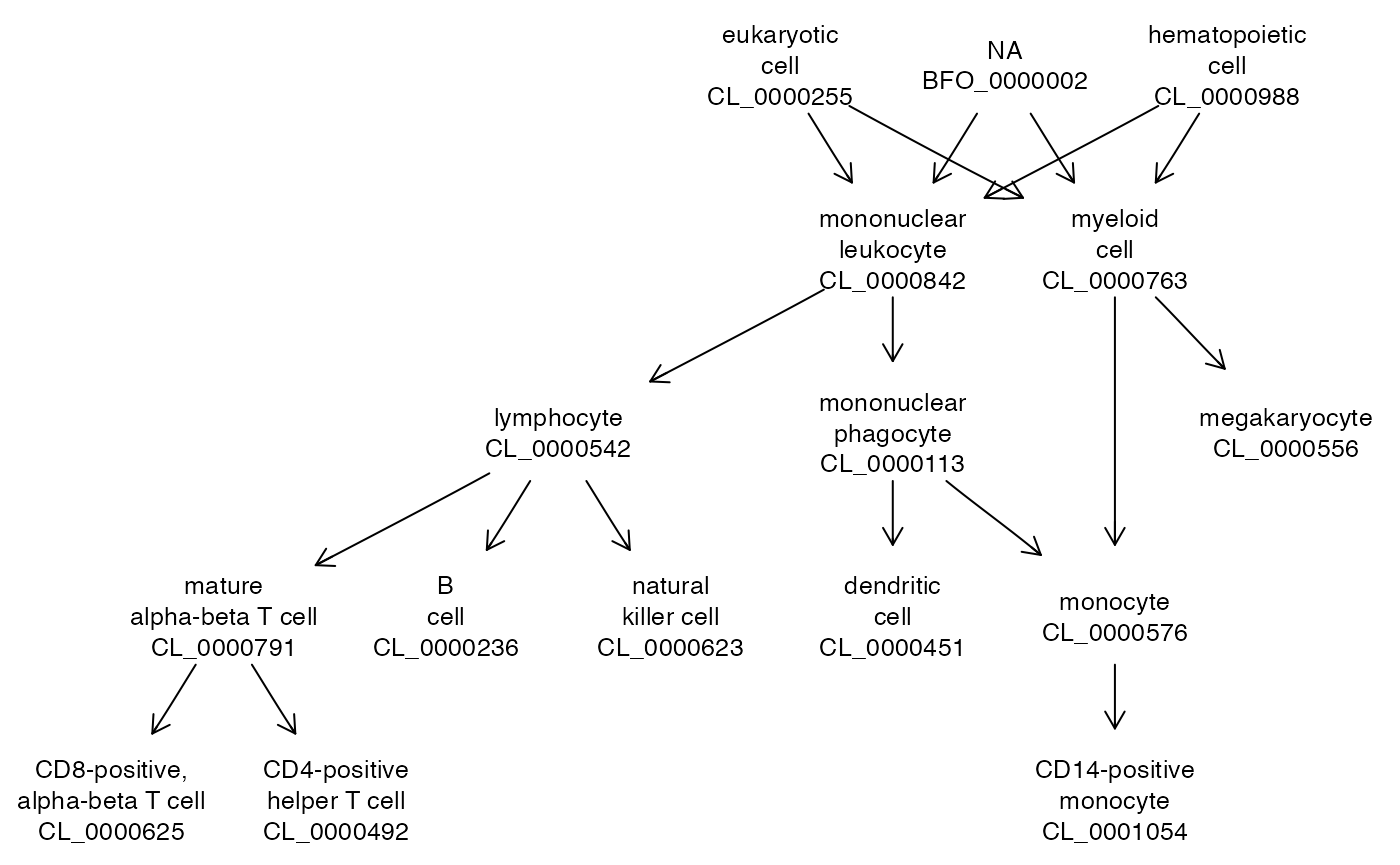
Illustrations with Human Phenotype ontology
We’ll obtain and ad hoc selection of 15 UBERON term names and visualize the hierarchy.
hpont_path = owl2cache(url="http://purl.obolibrary.org/obo/hp.owl")## resource BFC6 already in cache from http://purl.obolibrary.org/obo/hp.owl
hpents = setup_entities(hpont_path)
kp = grep("UBER", hpents$clnames, value=TRUE)[21:35]
plot(hpents, kp)## some index values not matched, ignoring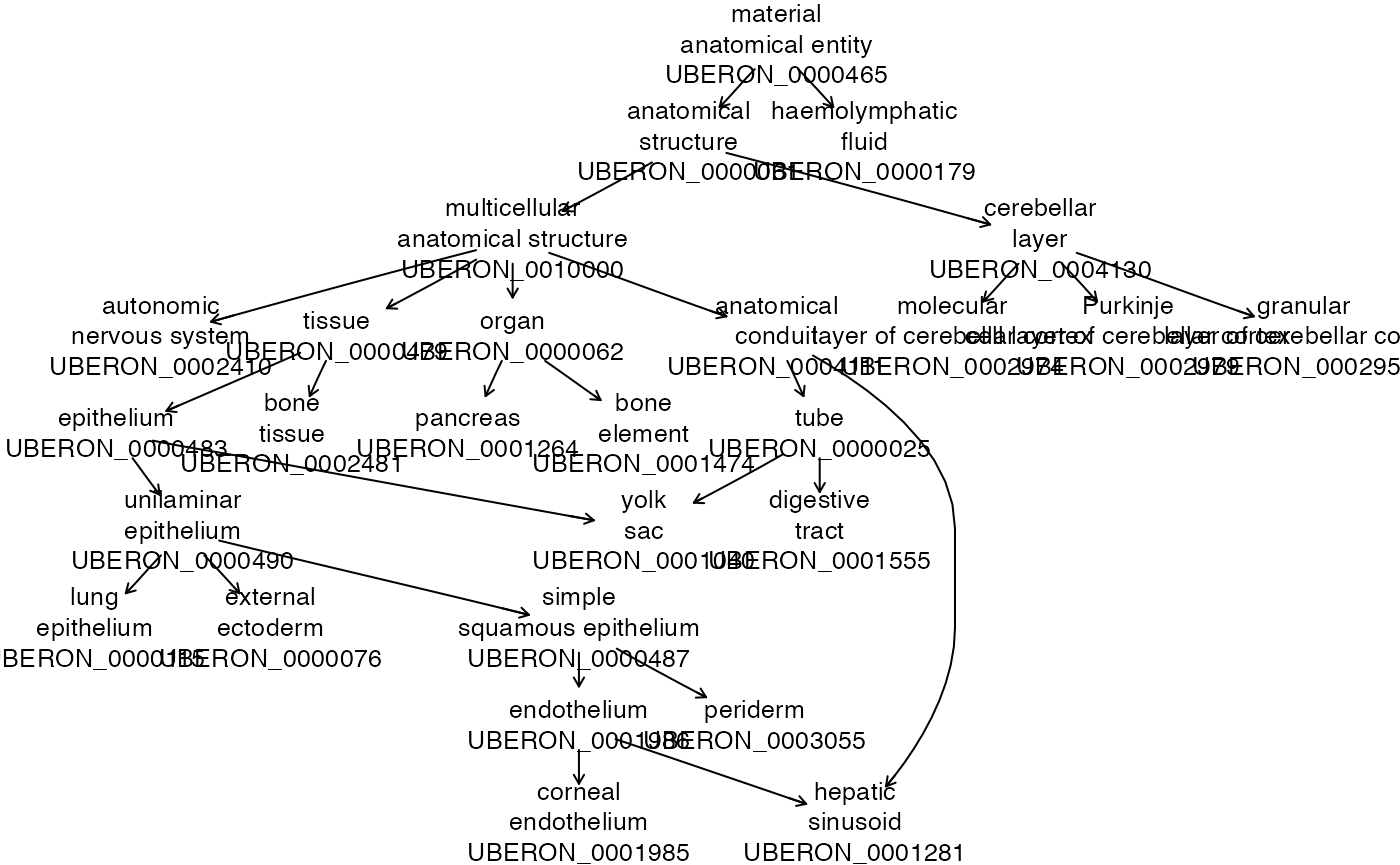
The prefixes of class names in the ontology give a sense of its scope.
##
## [,1]
## CHEBI 1848
## CL 1188
## GO 2518
## HP 19650
## HsapDv 12
## MPATH 75
## NBO 64
## PATO 569
## PR 206
## RO 1
## UBERON 5633To characterize human phenotypes ontologically, CL, GO, CHEBI, and UBERON play significant roles.
Caching
We described owl2cache above. By default, BiocFileCache’s cache is used. Query it for all available OWL files.
library(BiocFileCache)## Loading required package: dbplyr
ca = BiocFileCache()
bfcquery(ca, "owl")## # A tibble: 8 × 10
## rid rname create_time access_time rpath rtype fpath last_modified_time etag
## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 BFC1 aeo.… 2025-08-06… 2025-08-06… /Use… web http… 2020-04-14 07:31:… e70b…
## 2 BFC2 57b0… 2025-08-06… 2025-08-06… /Use… rela… /var… NA NA
## 3 BFC3 uber… 2025-08-06… 2025-08-06… /Use… web http… 2025-05-28 15:54:… 0x8D…
## 4 BFC6 hp.o… 2025-08-06… 2025-08-06… /Use… web http… 2025-05-06 15:39:… 0x8D…
## 5 BFC7 57b0… 2025-08-06… 2025-08-06… /Use… rela… /var… NA NA
## 6 BFC8 EDAM… 2025-08-08… 2025-08-08… /Use… web http… 2024-10-31 18:43:… 3171…
## 7 BFC9 11c5… 2025-08-08… 2025-08-08… /Use… rela… /var… NA NA
## 8 BFC11 cl.o… 2025-08-10… 2025-08-10… /Use… web http… 2025-07-30 12:34:… 0x8D…
## # ℹ 1 more variable: expires <dbl>At present there is no attempt to identify ontology version. This enhancement may be introduced in a future revision of ontoProc.
Once we have the path, setup_entities will deserialize the content to the owlents class.
Ontology of GWAS outcomes in the GWAS catalog
In this section we examine a package that is under development, gwasCatSearch. This uses ideas from the semantic sql project to improve performance of the most common operations on ontologies.
We’ll use the search app at shinyapps.io to explore connections between the GWAS catalog and the Experimental Factor Ontology used to annotate GWAS outcomes. Intermediate components are corpustools for enhancing discovery in natural language formulations of phenotypes, and SQLite, with tables representing ontology terms and their ancestors and descendants.
This segment gives some sense of how outcomes can be sorted according to the frequency of recorded hits in the catalog, and the associated EFO terms decoded.
library(gwasCatSearch)## Loading required package: corpustools##
## Attaching package: 'dplyr'## The following objects are masked from 'package:dbplyr':
##
## ident, sql## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
con = gwasCatSearch_dbconn()
dbListTables(con)## [1] "efo_dbxrefs" "efo_edges"
## [3] "efo_entailed_edges" "efo_labels"
## [5] "efo_synonyms" "gwascatalog_associations"
## [7] "gwascatalog_mappings" "gwascatalog_metadata"
## [9] "gwascatalog_references" "uberon_edges"
## [11] "uberon_entailed_edges" "uberon_labels"
## [13] "uberon_synonyms" "version_info"
tbl(con, "gwascatalog_associations") |> select(MAPPED_TRAIT_URI) |>
group_by(MAPPED_TRAIT_URI) |> summarise(n=n()) |> arrange(desc(n))## # Source: SQL [?? x 2]
## # Database: sqlite 3.50.3 [/private/var/folders/yw/gfhgh7k565v9w83x_k764wbc0000gp/T/RtmpfK2EoK/gwascatalog_search.db]
## # Ordered by: desc(n)
## MAPPED_TRAIT_URI n
## <chr> <int>
## 1 http://www.ebi.ac.uk/efo/EFO_0004339 25475
## 2 http://www.ebi.ac.uk/efo/EFO_0004340 13668
## 3 http://www.ebi.ac.uk/efo/EFO_0004612 8100
## 4 http://www.ebi.ac.uk/efo/EFO_0007937 8045
## 5 http://www.ebi.ac.uk/efo/EFO_0007788 7796
## 6 http://www.ebi.ac.uk/efo/EFO_0004530 7749
## 7 http://www.ebi.ac.uk/efo/EFO_0005664 6815
## 8 http://www.ebi.ac.uk/efo/EFO_0004747 6758
## 9 http://www.ebi.ac.uk/efo/EFO_0004611 6568
## 10 http://www.ebi.ac.uk/efo/EFO_0007874 6547
## # ℹ more rows## # Source: SQL [?? x 6]
## # Database: sqlite 3.50.3 [/private/var/folders/yw/gfhgh7k565v9w83x_k764wbc0000gp/T/RtmpfK2EoK/gwascatalog_search.db]
## Subject Object IRI DiseaseLocation Direct Inherited
## <chr> <chr> <chr> <chr> <int> <int>
## 1 EFO:0004340 body mass index http… NA 307 31
## 2 EFO:0004612 high density lipoprotein c… http… NA 489 17