owlents: using OWL directly in ontoProc
Vincent J. Carey, stvjc at channing.harvard.edu
June 22, 2025
Source:vignettes/owlents.Rmd
owlents.RmdIntroduction
In Bioconductor 3.19, ontoProc can work with OWL RDF/XML serializations of ontologies, via the owlready2 python modules.
The owl2cache function retrieves OWL from a URL or file and places it in a cache to avoid repetitious retrievals. The default cache is the one defined by BiocFileCache::BiocFileCache(). Here we work with the cell ontology. setup_entities2 will use basilisk to acquire owlready2 python modules that parse the OWL and produce an ontology_index instance (defined in CRAN package ontologyIndex).
library(ontoProc)
clont_path = owl2cache(url="http://purl.obolibrary.org/obo/cl.owl")
cle = setup_entities2(clont_path)
cle## Ontology with 17107 terms
##
## Properties:
## id: character
## name: character
## parents: list
## children: list
## ancestors: list
## obsolete: logical
## Roots:
## BFO_0000002 - continuant
## BFO_0000003 - occurrent
## SO_0000704 - NA
## SO_0001260 - sequence_collection
## CHEBI_18059 - lipid
## CHEBI_25905 - peptide hormone
## CHEBI_16646 - carbohydrate
## CHEBI_33696 - nucleic acid
## CHEBI_63299 - carbohydrate derivative
## CHEBI_33694 - biomacromolecule
## ... 333 moreThe usual plotting approach works.
sel = c("CL_0000492", "CL_0001054", "CL_0000236",
"CL_0000625", "CL_0000576",
"CL_0000623", "CL_0000451", "CL_0000556")
onto_plot2(cle, sel)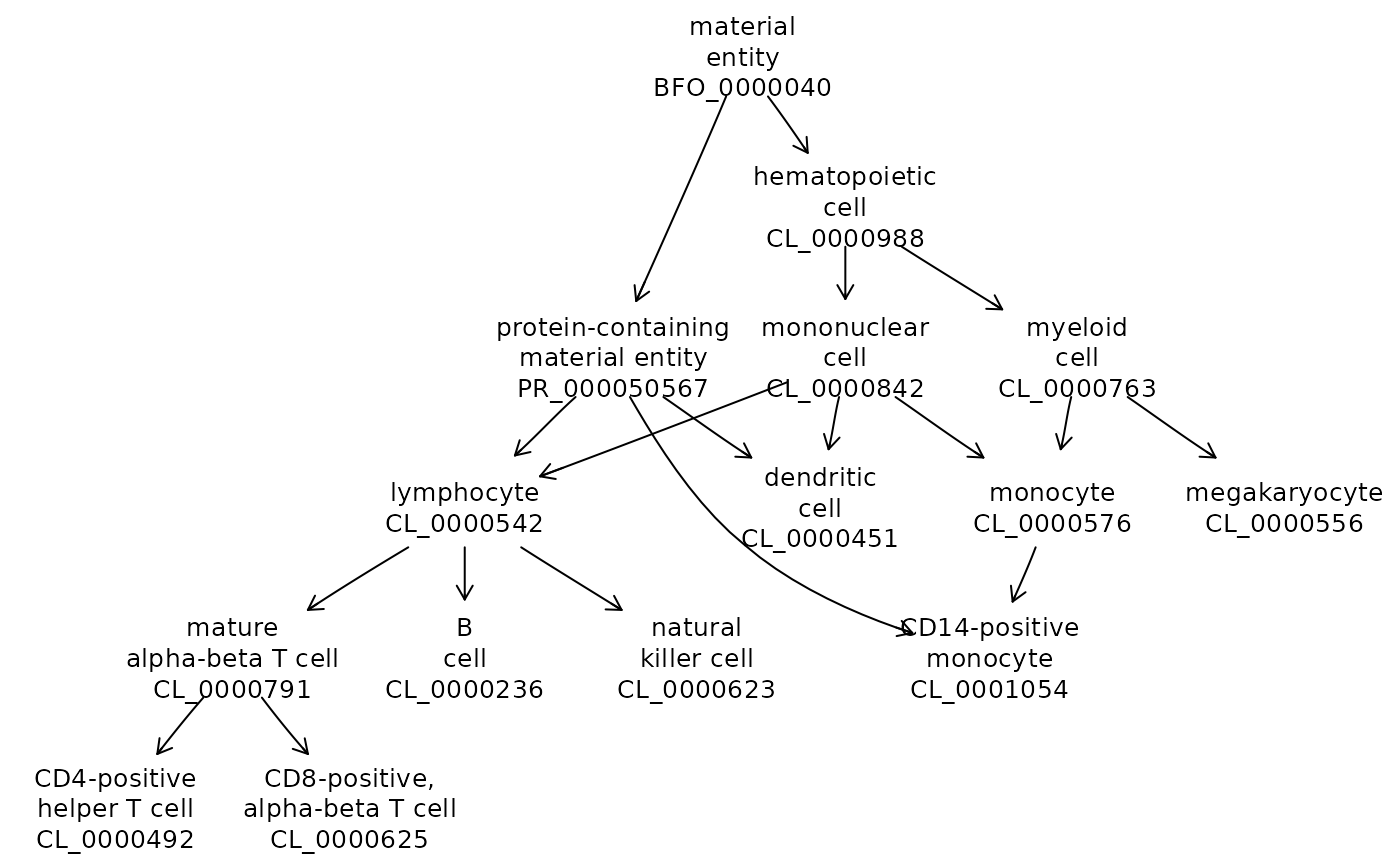
Illustration with Human Phenotype ontology
We’ll obtain and ad hoc selection of 15 UBERON term names and visualize the hierarchy.
hpont_path = owl2cache(url="http://purl.obolibrary.org/obo/hp.owl")## resource BFC1019 already in cache from http://purl.obolibrary.org/obo/hp.owl
hpents = setup_entities2(hpont_path)
kp = grep("UBER", names(hpents$name), value=TRUE)[21:30]
onto_plot2(hpents, kp)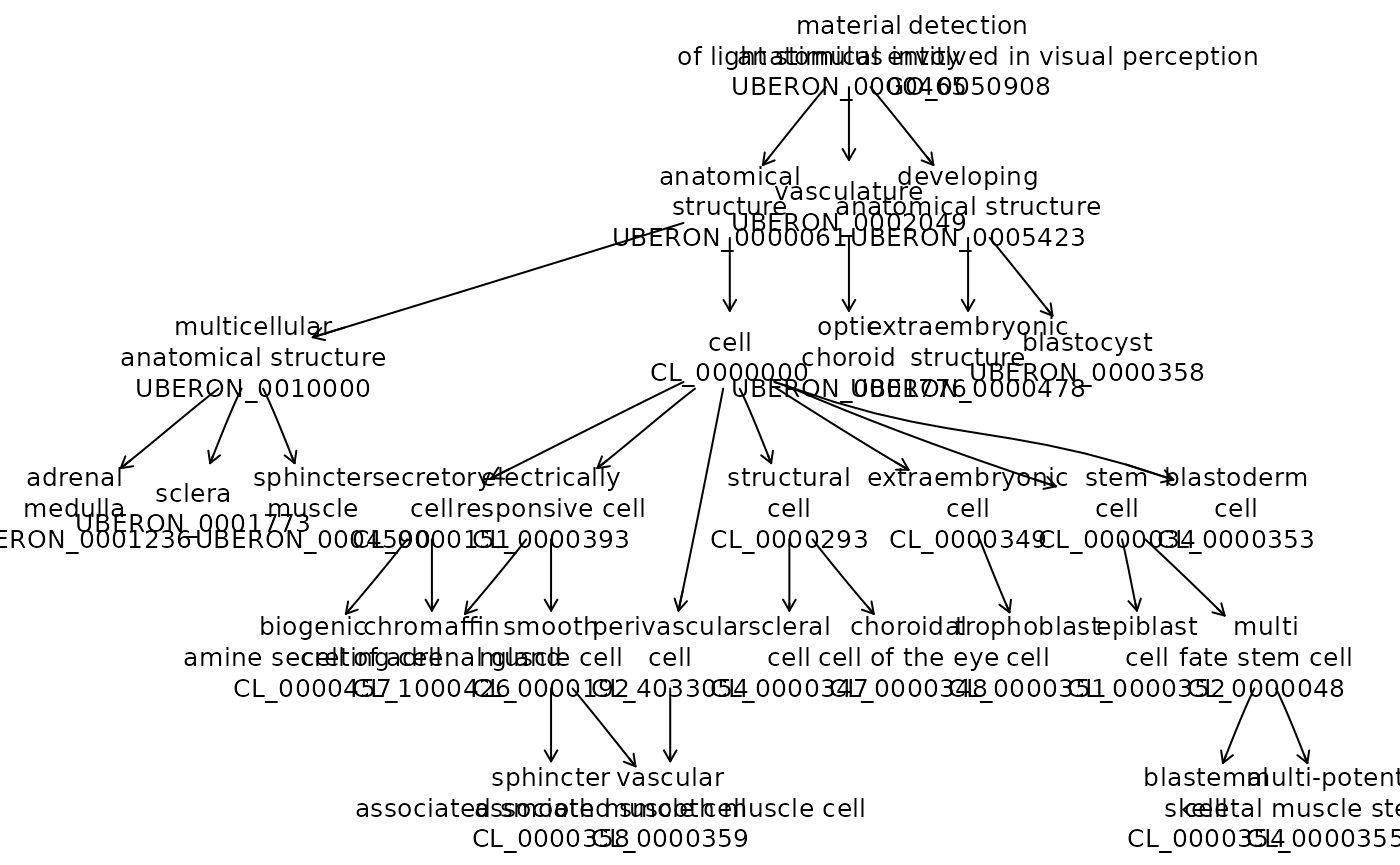
The prefixes of class names in the ontology give a sense of its scope.
##
## [,1]
## BFO 14
## CHEBI 1595
## CL 1142
## GO 2679
## HP 18252
## HsapDv 12
## MPATH 75
## NBO 143
## PATO 567
## PR 205
## RO 1
## UBERON 5602To characterize human phenotypes ontologically, CL, GO, CHEBI, and UBERON play significant roles.