produce a heatmap from a specialized CITE-seq SingleCellExperiment
Source:R/adt_profiles.R
adtProfiles.Rdproduce a heatmap from a specialized CITE-seq SingleCellExperiment
adtProfiles(x, lb = -3, ub = 3, do_z = FALSE)Arguments
- x
SingleCellExperiment instance that has an `se.averaged` component in its metadata
- lb
numeric(1) lower bound on 'breaks' sequence for ComplexHeatmap::pheatmap, defaults to -3
- ub
numeric(1) upper bound on 'breaks' sequence for ComplexHeatmap::pheatmap, defaults to 3
- do_z
logical(1) if TRUE, divide the residuals by their standard deviation across clusters, defaults to false
Value
ComplexHeatmap::pheatmap instance
side effect of pheatmap::pheatmap call
Note
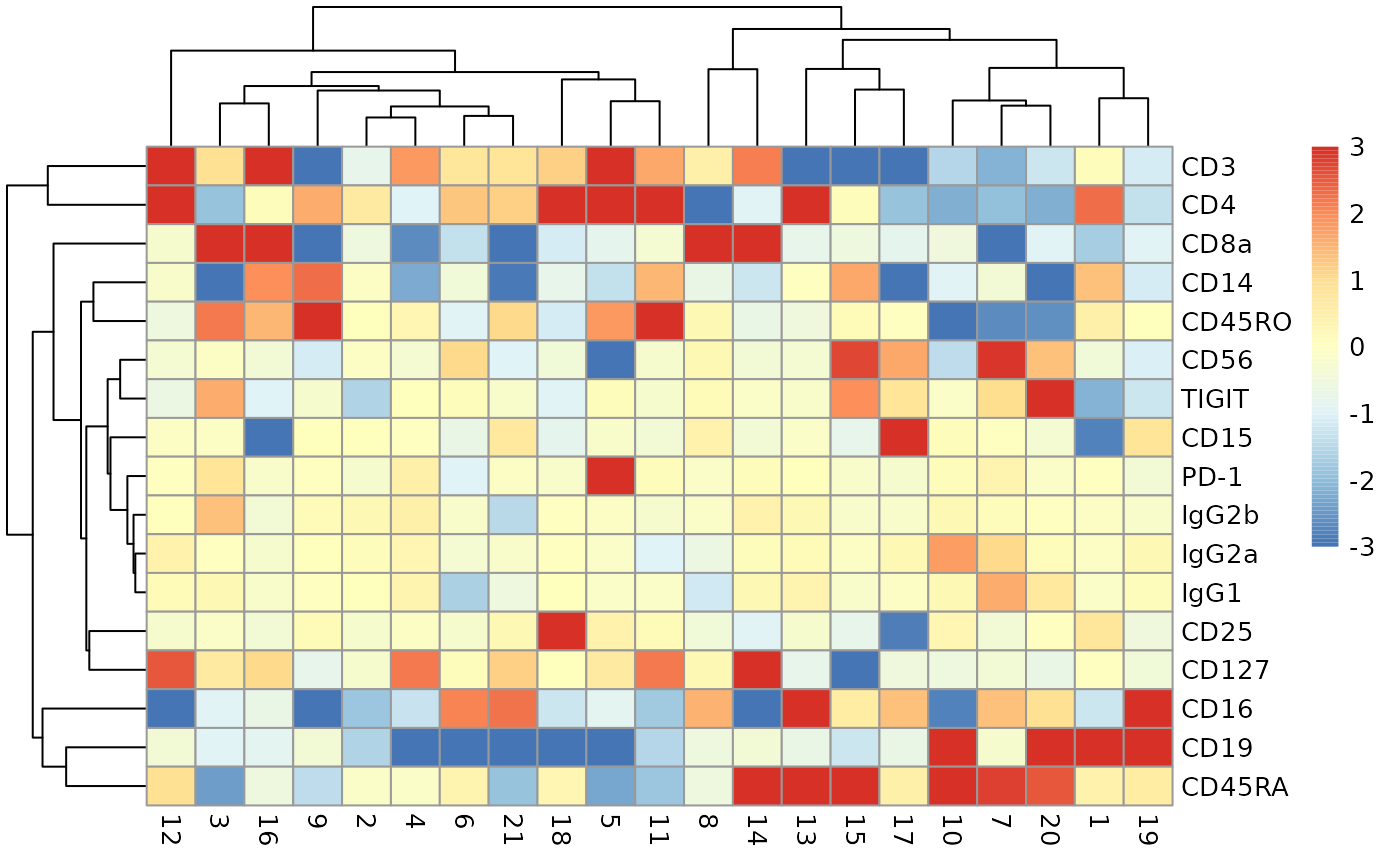
See the OSCA book ch12.5.2 for the application.
Examples
ch12sce <- getCh12Sce()
adtProfiles(ch12sce)
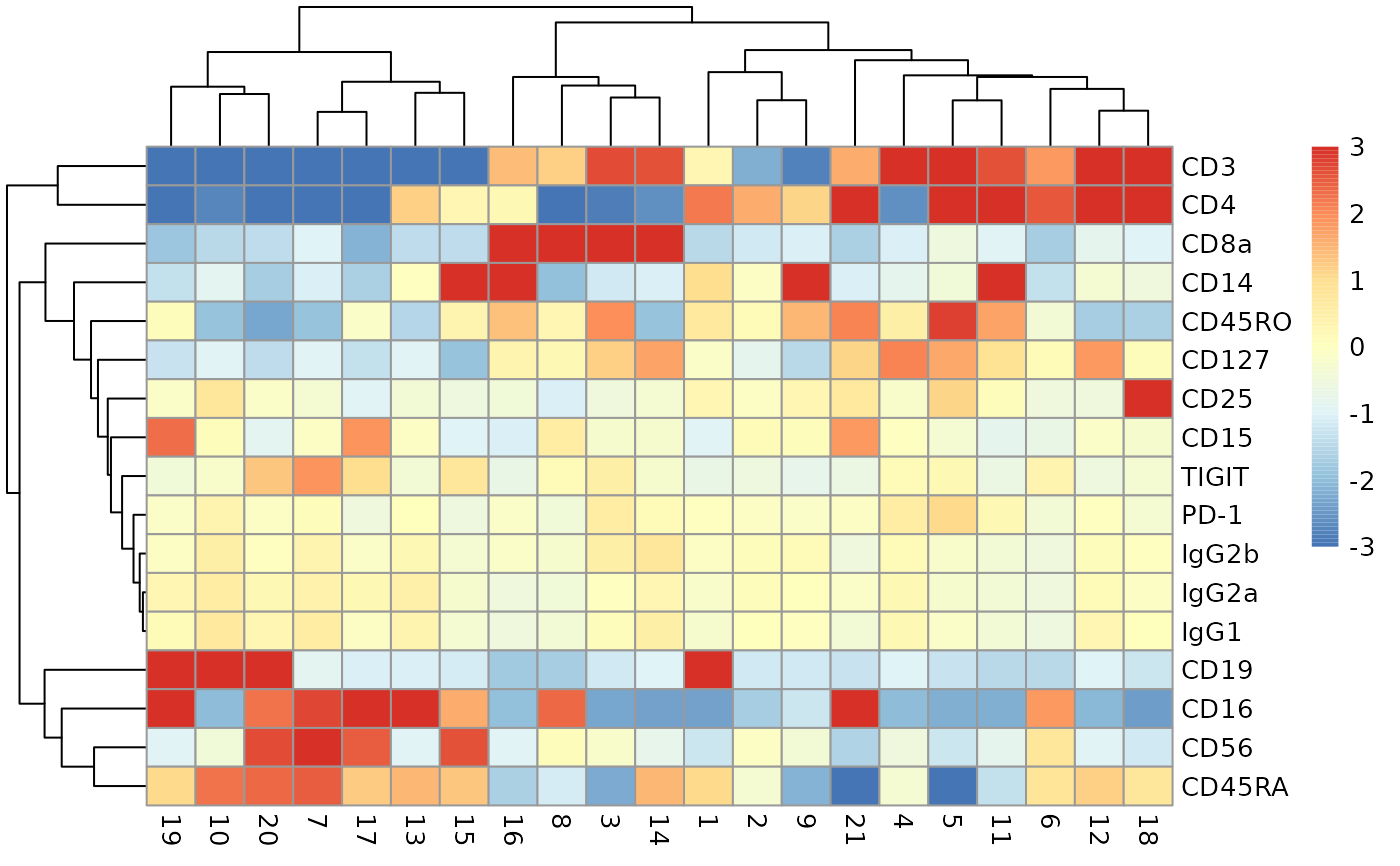 adtProfiles(ch12sce, do_z = TRUE)
adtProfiles(ch12sce, do_z = TRUE)