S6 HCA, ontologies, shiny
Vincent J. Carey, stvjc at channing.harvard.edu
January 09, 2025
Source:vignettes/S6_hca_onto.Rmd
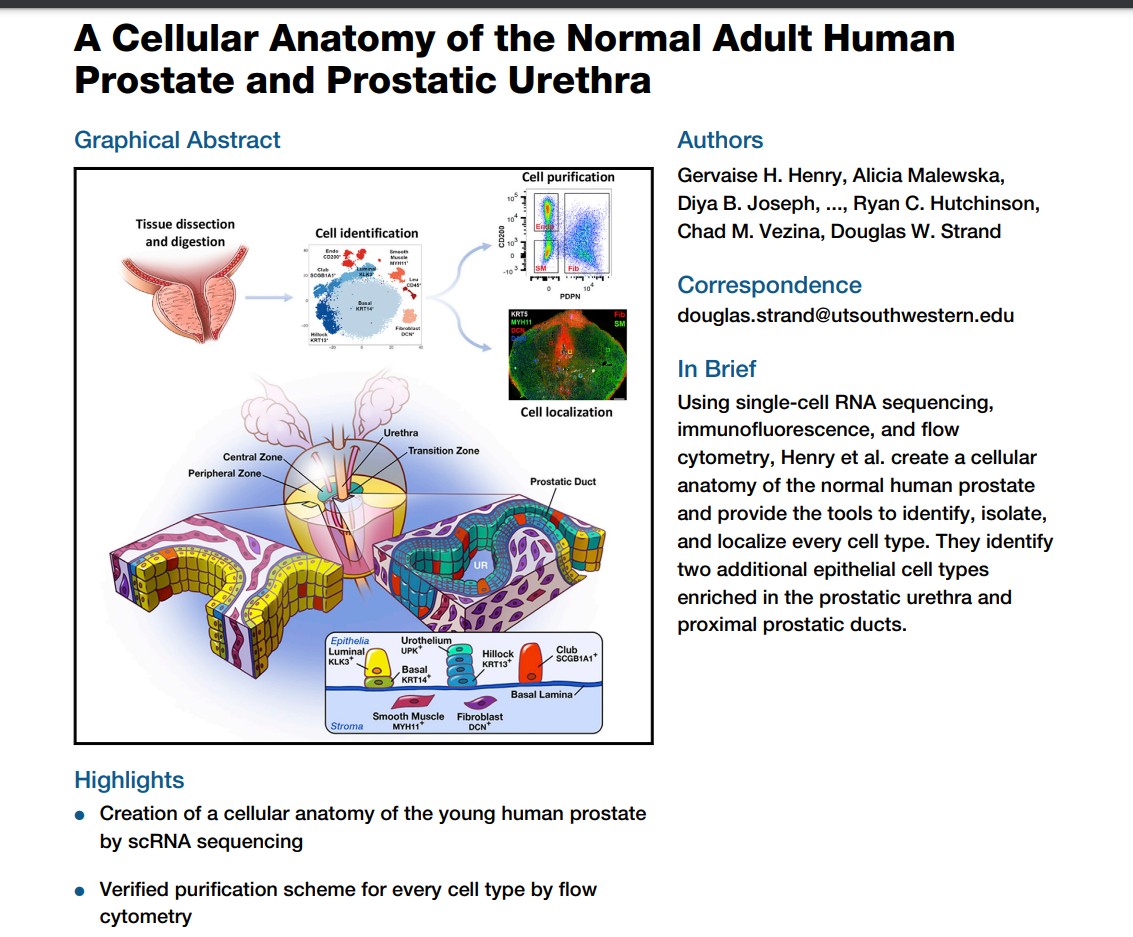
S6_hca_onto.RmdA paper on the atlas of the prostate
overview
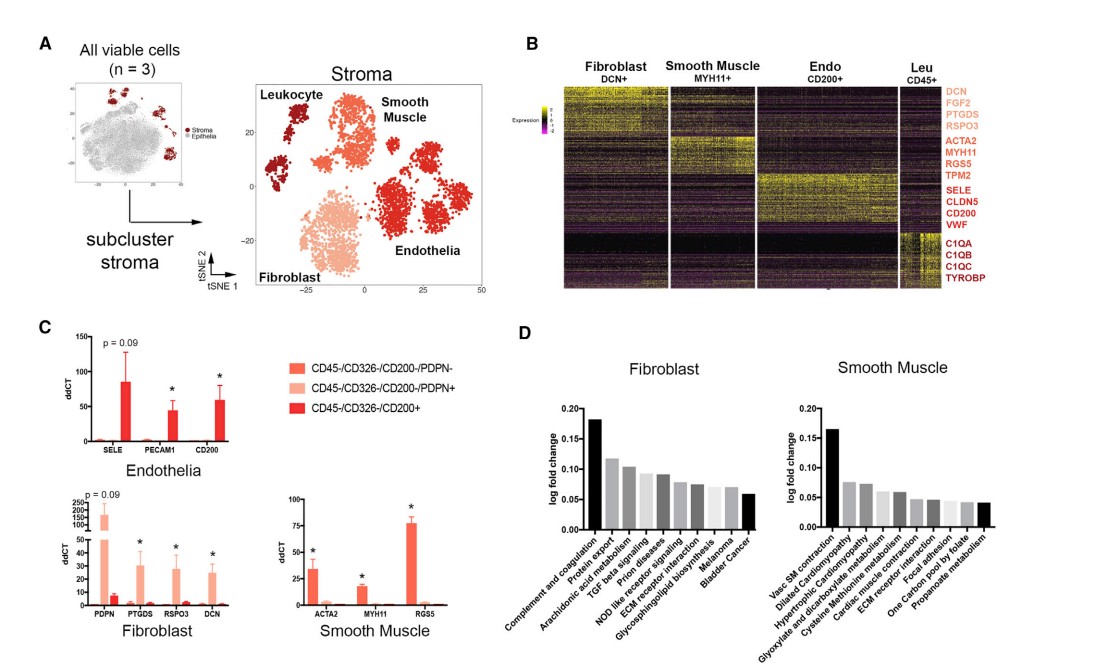
normals
The hca package
Picking a project; Enumerating and downloading loom files
projectId = "53c53cd4-8127-4e12-bc7f-8fe1610a715c"
file_filter <- filters(
projectId = list(is = projectId),
fileFormat = list(is = "loom")
)
pfile = files(file_filter)
pfile$projectTitle[1]## [1] "A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra"
#pfile |> files_download()Working with loom
Very superficial filtering (to 60000 cells) and development of PCA
library(LoomExperiment)
f1 = import("/home/stvjc/.cache/R/hca/36e582f7c6e_36e582f7c6e.loom")
f1
names(colData(f1))
library(scater)
sf1 = as(f1, "SingleCellExperiment")
sf1
library(scuttle)
assay(sf1[1:4,1:4])
assayNames(sf1) = "counts"
litsf1 = sf1[,1:60000]
z = DelayedArray::rowSums(assay(litsf1))
mean(z==0)
todrop = which(z==0)
litsf2 = litsf1[-todrop,]
assay(litsf2)
litsf2 = logNormCounts(litsf2)
litsf2 = runPCA(litsf2)This code is blocked until a bucket is made available with the filtered data.
library(SingleCellExperiment)
if (!exists("litsf2")) load("litsf2.rda") # run code above, must have HDF5 in cache
metadata(litsf2)> str(litsf2) # 22MB on disk (no quantifications)
Formal class 'DelayedMatrix' [package "DelayedArray"] with 1 slot
..@ seed:Formal class 'DelayedAperm' [package "DelayedArray"] with 2 slots
.. .. ..@ perm: int [1:2] 2 1
.. .. ..@ seed:Formal class 'DelayedSubset' [package "DelayedArray"] with 2 slots
.. .. .. .. ..@ index:List of 2
.. .. .. .. .. ..$ : int [1:60000] 1 2 3 4 5 6 7 8 9 10 ...
.. .. .. .. .. ..$ : int [1:23420] 13 20 22 23 31 33 34 35 36 37 ...
.. .. .. .. ..@ seed :Formal class 'HDF5ArraySeed' [package "HDF5Array"] with 7 slots
.. .. .. .. .. .. ..@ filepath : chr "/home/stvjc/.cache/R/hca/36e582f7c6e_36e582f7c6e.loom"
.. .. .. .. .. .. ..@ name : chr "/matrix"
.. .. .. .. .. .. ..@ as_sparse: logi FALSE
.. .. .. .. .. .. ..@ type : chr NA
.. .. .. .. .. .. ..@ dim : int [1:2] 382197 58347
.. .. .. .. .. .. ..@ chunkdim : int [1:2] 64 64
.. .. .. .. .. .. ..@ first_val: int 0stvjc@stvjc-XPS-13-9300:~/CSAMA_HCA$ ls -tl /home/stvjc/.cache/R/hca/36e582f7c6e_36e582f7c6e.loom
-rw-rw-r-- 1 stvjc stvjc 1206062245 Jun 21 22:37 /home/stvjc/.cache/R/hca/36e582f7c6e_36e582f7c6e.loomOntologies, EBI OLS, rols (thanks Laurent Gatto!), ontoProc::ctmarks
Definition: from Wikipedia
In computer science and information science, an ontology encompasses a representation, formal naming, and definition of the categories, properties, and relations between the concepts, data, and entities that substantiate one, many, or all domains of discourse. More simply, an ontology is a way of showing the properties of a subject area and how they are related, by defining a set of concepts and categories that represent the subject.
Every academic discipline or field creates ontologies to limit complexity and organize data into information and knowledge. Each uses ontological assumptions to frame explicit theories, research and applications. New ontologies may improve problem solving within that domain. Translating research papers within every field is a problem made easier when experts from different countries maintain a controlled vocabulary of jargon between each of their languages.
Applications in genomics
- Gene Ontology: Genes and gene products in subdomains of BP, MF, CC – biological process, molecular function, cellular component
- Human Phenotype Ontology
- UBERON - cross-species anatomy
- Cell ontology
- Cell line ontology
- EFO - experimental factor ontology
Tags from any of these can be encountered in various annotation resources.
rols: Basic idea
- OLS is ontology lookup service
- has API
- rols package help to interrogate the service
- ontologies are everywhere
Learn about ‘smooth muscle’ with rols
## Object of class 'OlsSearch':
## query: smooth muscle
## requested: 100 (out of 58452)
## response(s): 0ontoProc – capitalizing on ontologyIndex (thanks Daniel Greene!), Rgraphviz (thanks Kasper Hansen!)
## loading from cache
head(co$name)## BFO:0000002 BFO:0000003 BFO:0000004
## "continuant" "occurrent" "independent continuant"
## BFO:0000006 BFO:0000015 BFO:0000016
## "spatial region" "process" "disposition"The ctmarks app: walk through linked ontologies such as PR and present additional facets about the concept in focus
Limitation: the OBO representation in use is outdated and out-links are sparse
chk = ctmarks(co)Projects: - use rols to get more interesting information about terms into the app - update the ontology resources - go beyond OBO … but not all the way to OWL? Evaluate the UI/UX needed to broaden ontology usage - impacts: data integration, precision of annotation, cognitive efficiency