A2 Acquiring data on COVID-19 using downloads and ‘APIs’
Vincent J. Carey, stvjc at channing.harvard.edu
January 07, 2023
Source:vignettes/A2downloads.Rmd
A2downloads.RmdIntroduction
The teachCovidData package includes snapshots of data as a convenience to students and teachers.
Because the pandemic is ongoing, “up-to-date” data are of considerable interest.
- Up-to-date records are important for identifying acute developments.
- Unless strong real-time quality control procedures are in place, up-to-date records may include misleading errors that will take time to correct.
Using APIs: formats and ‘pagination’
Application Programming Interfaces (APIs) specify how we can query resources on the web to obtain data. Usually it is sufficient to compose a URL and use the URL with a “client” to trigger delivery of data. We will demonstrate this in this section.
Format options
The CDC provides APIs to COVID-related data using products of a company called “Tyler”. Their documentation of API usage and data formats can be found here.
Heres’ a snapshot of the documentation about data formats and API elements:
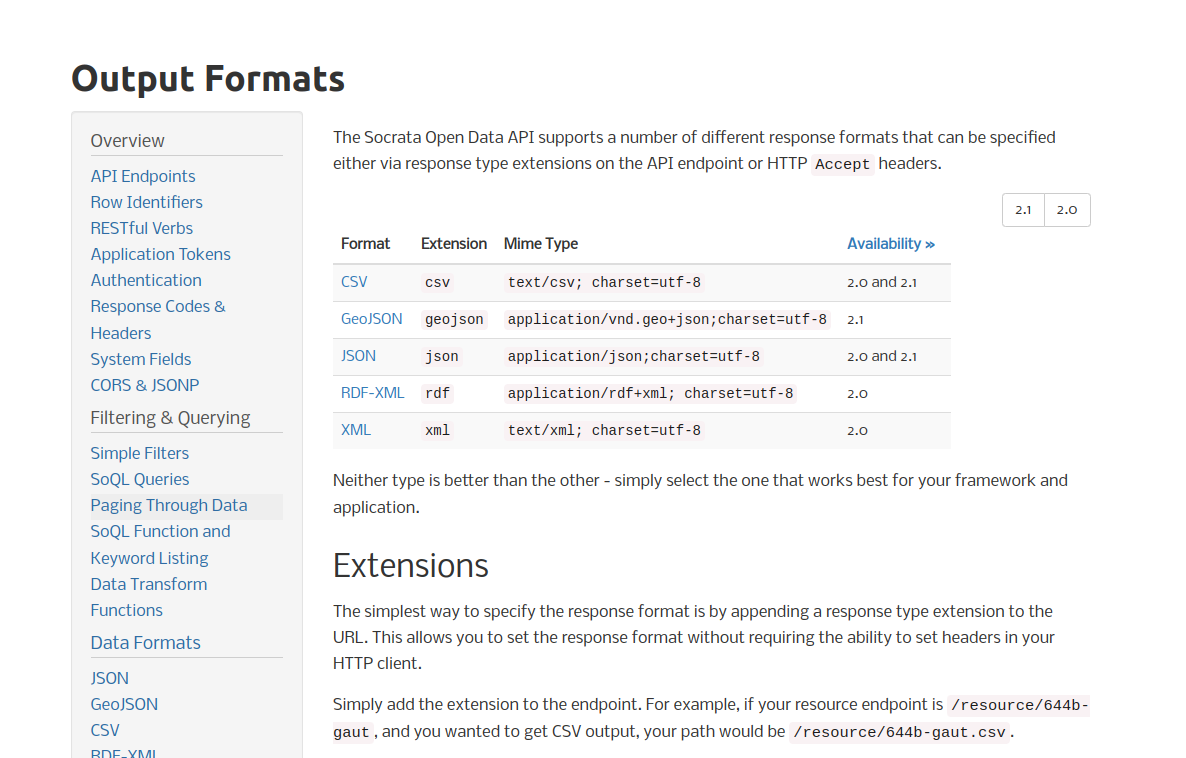
format snapshot
Notice the ‘2.1|2.0’ in the upper right corner. This is indicative of availability of different versions of the API.
It is very important to understand the “road map” of a data provider or data consumption process. Information technology and data production methods are highly dynamic, and producers and consumers need to be able to adapt to changes.
Using an API in R; pagination
The “API” button at the CDC download site for vaccine data by jurisdiction produces a URL:
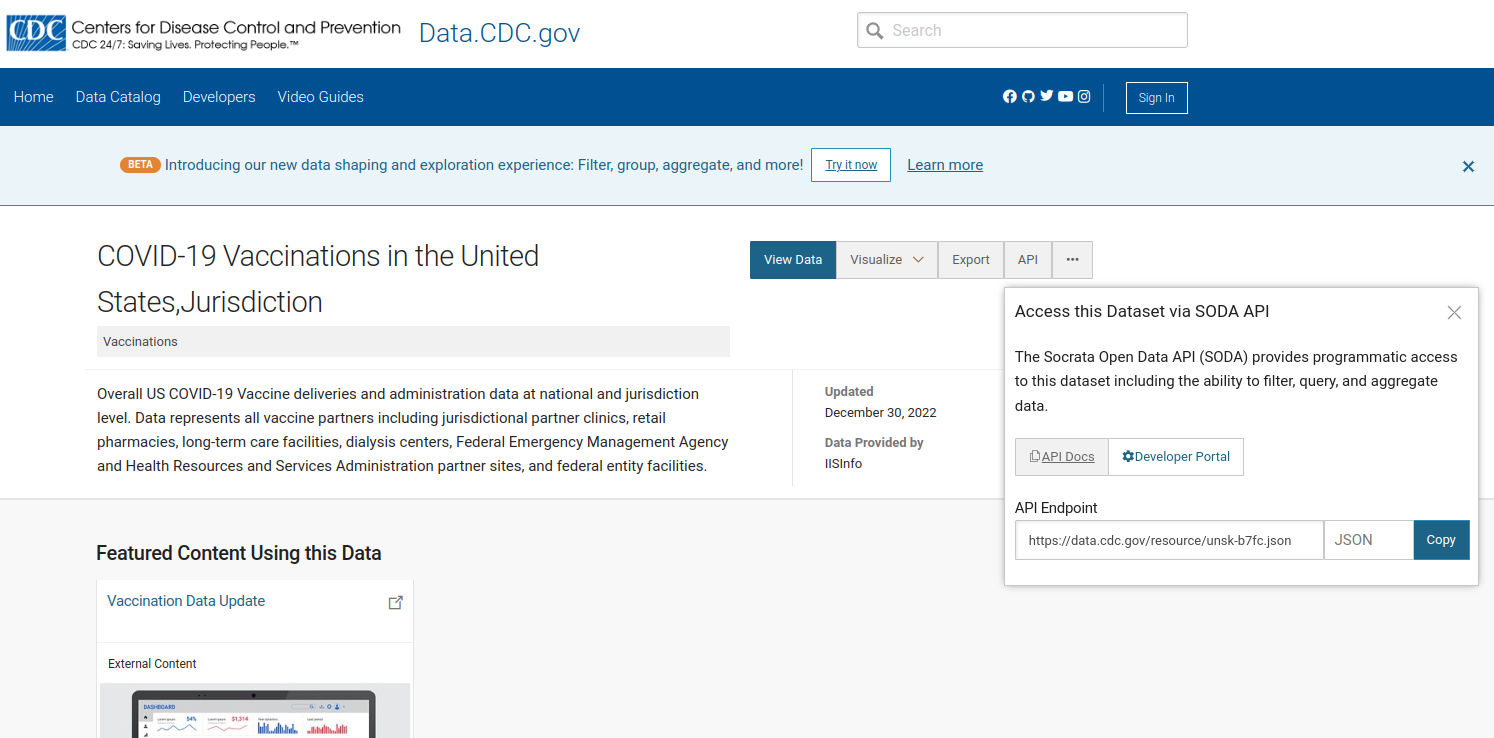
JSON endpoint
library(jsonlite)
dat = fromJSON("https://data.cdc.gov/resource/unsk-b7fc.json")
dim(dat) # fromJSON simplifies when possible## [1] 1000 109
datatable(dat[1:50,1:40]) # illustrative subsetWe have received a data.frame by retrieving JSON using the CDC data provider URL. The fact that dim(dat) returns exactly 1000 “rows” is a hint that we have received only one “page” of a larger resource. This concept is discussed in the API documentation.
We can reformulate the query to retrieve a larger collection of records.
## [1] 2000 109We have to be careful to avoid requesting unwieldy payloads from data providers. Modifications to URLs that support filtering of resources are defined in API documentation.
Using GitHub to distribute data
New York City’s Department of Health distributes data in a GitHub repository. The instructions suggest cloning the repository to work with the data.
For example information on variant prevalence is described here and instructions for use start in the README.md of the repository.